[Go back to the main documentation for matrix2png]
The first examples use this data file, which is part of the "Eisen" data set from Stanford.
matrix2png -data testdata.rdb -size 8:8 -map 1 -range -2:2 -numr 10 >! example1.png
About the options: -size determines the size of each feature, 8x8 pixels in this case. -map 1 selects the color scheme from a presets. -range -2:2 specifies that the ends of the color range correspond to values of -2 and 2. This means that values less than -2 and greater than 2 will be 'clipped' and displayed as the 'mincolor' and 'maxcolor' respectively. -numr 10 determines that only 10 rows of data will be shown.

Use the -r, -c, and -s options to get row labels, column labels, and a scalebar. The -trim 5 limits the color range to the middle 90% of the data values - the highest and lowest 5% are 'clipped'.
matrix2png -data testdata.rdb -size 8:8 -r -c -s -map 3 -trim 5 -numr 10 >! example2.png

This is example 2 without trimming.
matrix2png -data testdata.rdb -size 8:8 -map 3 -range -2:2 -numr 10 >! example3.png

You can (partly) select the colors you want to use instead of a map, using -mincolor and -maxcolor. The use of -b makes the color range go through black in the middle. Use -e to get ellipses. The low contrast in this image comes from the failure to use -range or -trim or -con.
matrix2png -data testdata.rdb -size 8:8 -mincolor red -maxcolor green -e -b -bkgcolor black >! example4.png
Note that similar results can be obtained with -map 3 instead of specifying -mincolor, -maxcolor, and -b.

The -bkgcolor option sets the background color. This is preset map number 4.
matrix2png -data testdata.rdb -size 8:8 -map 4 -e -range -2:2 -bkgcolor black -numr 10 >! example5.png

Add dividers between each block with -d.
matrix2png -data testdata.rdb -size 8:8 -map 1 -d -range -2:2 -numr 10 >! example6.png
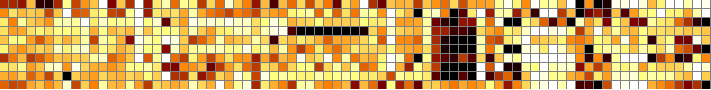
This one is of data that is categorical, not continuous, from a data file representing a gene structure prediction compared to a reference structure. It uses a map file, NM_000041.map.
matrix2png -size 1:16 -s -r -dmap NM_000041.map -data NM_000041.ds.mtx >! NM_000041.10.png

The following are additional examples of images created with matrix2png in diverse contexts. Because these images are larger than the cases above, you have to follow the link to the image. You can also follow the links to the data files; be warned that some of them are quite large.
Each entry is a Boolean indicating whether the two genes interact, based on a recently published study (Von Mering, et al.). Only a portion of the data file is shown, using the -numc and -numr options.
matrix2png -data vonmering.count5.reduced.mtx -map 15 -size 8:8 -rdc -numc 100 -numr 100 > vonmering.count5.reduced.png
This diagram shows the dynamic programming matrix produced by an automatic gene finding algorithm. Each row in the matrix corresponds to a state in the gene finding hidden Markov model, and each column corresponds to a sequence position. The codon structure is apparent in the many rows that have repetitions with period three.
matrix2png -data HSG0S2A.post-decode.numcopies1.stats.mtx.txt -src -size 8:8 -range -100:0 > HSG0S2A.post-decode.numcopies1.stats.png
Each entry in this matrix is the log E-value of the BLAST score between two protein domains in the SCOP database.
matrix2png -data scop.1.53.1e-25.blast.matrix -numr 200 -numc 200 -trim 1 -size 8:8 -rcs -mincolor 20:20:20 -maxcolor red > scop.1.53.1e-25.blast.trim.png
This is a schematic diagram of a multiple alignment of a family of SCOP protein domains. In the figure, insertions and deletions are white, and most amino acids are grey. The amino acids that are black correspond to subsequences that are useful in recognizing occurrences of this domain, as determined by mismatch spectrum support vector machine.
matrix2png -data 1.34.1.13Ad1cb1__.mtx.txt -numr 200 -size 4:4 -mincolor white -maxcolor black > 1.34.1.13Ad1cb1__.png